Events
News
Zhanxin Wang Laboratory in Beijing Normal University and collaborators published a Nature article revealing the molecular basis of nucleosome engagement and H3K36 methylation by the NSD family of histone lysine methyltransferases
On Dec. 24 2020, Nature published the article entitled “Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases” from the collaborated results of the Zhanxin Wang laboratory in the College of Life Sciences Beijing Normal University, the cryo-EM center in the Southern University of Science and Technology, the Dinshaw Patel laboratory in Memorial Sloan-Kettering Cancer Center, and the Or Gozani laboratory in Stanford University, which for the first time reported the catalytic mechanism of NSD proteins at the nucleosome level, and the impact of NSD protein mutations on human cancer.
Human NSD proteins include NSD1, NSD2 and NSD3 three members, all of which catalyze the dimethylation on Lys36 of histone H3. NSD proteins are key epigenetic regulators that regulate chromatin structure and play crucial roles in higher eukaryotes. NSD mutations are closely related to human cancers. For example, NSD3 is related to acute myeloid leukemia (AML) and head and neck cancer, NSD2 is related to multiple myeloma and the Wolf-Hirschhorn Syndrome. However, currently no effective drugs are available for this family of proteins. This work solved high resolution cryo-EM structures of NSD2 and NSD3 with bound nucleosomes, revealed that NSD2 and NSD3 tended to bind nucleosomes with linker DNAs. NSD2 or NSD3 unwrapped the nucleosomal DNA at the entry-exit sites, which facilitates the insertion of its catalytic region in-between the unwrapped DNA and the core histones. NSD proteins contact the nucleosome through a series of DNA- and histone-specific interactions, which enable them to exhibit an extended conformation on the nucleosomes, thus getting activated to exert their H3K36 specific methyltransferase activity. This work further found that several cancer-related mutations, such as E1099K and T1150A mutations in NSD2, and their corresponding E1181K and T1232A mutations in NSD3, increase the catalytic activity of NSD2 or NSD3, and increased the cell proliferation rates. In addition, these hyper-active mutations of NSD3 will increase the tumor growth rate in nude mice. Structural analysis revealed that although the E1099K mutation is away from the catalytic site, it will increase the catalytic activity of NSD2 through enhancing its interaction with the nucleosome DNA. T1232A mutation is close to the catalytic site of NSD3, this mutation increased the catalytic activity of NSD3 possibly through stabilizing the interaction between NSD3 and the H3 tail. Overall, this work for the first time revealed the catalytic mechanism of NSD proteins on nucleosomes, which facilitates our understanding of the physiology function of NSD proteins, and provides structural basis for their drug development.
This work is the collaborated results of the Zhanxin Wang laboratory from Beijing Normal University, the cryo-EM center of the Southern University of the Science and Technology (SUSTech), the Dinshaw Patel laboratory from the Memorial Sloan-Kettering Cancer Center, and the Or Gozani laboratory from Stanford University. Wanqiu Li from SUSTech, Wei Tian and Pujuan Deng from Zhanxin Wang’s lab, Gang Yuan from Or Gozani’s lab are co-first authors. Zhanxin Wang, Dinshaw Patel and Or Gozani are corresponding authors. Guohong Li from the Institute of Biophysics, CAS, Ning Gao from Peking University and Xin Gong from SUSTech helped on this work. This work was supported by the National Natural Science Foundation of China (NSFC), Fundamental Research Funds for the Central Universities, funds from SUSTech and the Shenzhen city, and funds from US. The cryo-EM data were collected from the cryo-EM centers from SUSTech and the National Center for Protein Science Shanghai (NCPSS).
Online link: https://www.nature.com/articles/s41586-020-03069-8
Figure of the title and authors:

Figure:
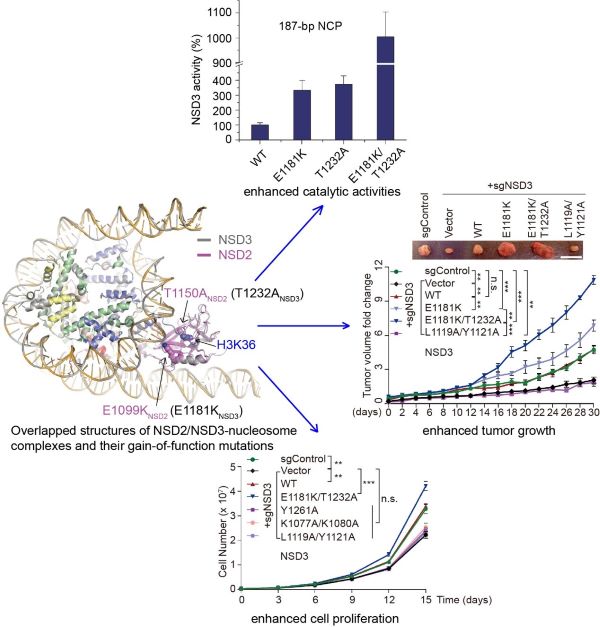
Figure legend: Overlapped structures of NSD2 and NSD3 with bound nucleosomes. Some cancer related mutations enhanced the catalytic activity of NSD proteins, enhanced the cell proliferation rates and enhanced the tumor growth rates in nude mice.


